Note
Click here to download the full example code
Bayesian Independent Component Analysis¶
Out:
2
# Author: Pierre Ablin <pierre.ablin@ens.fr>
#
# License: MIT
import torch
from ksddescent import ksdd_lbfgs
from ksddescent.contenders import svgd
import matplotlib.pyplot as plt
import numpy as np
def amari_distance(W, A):
"""
Computes the Amari distance between two matrices W and A.
It cancels when WA is a permutation and scale matrix.
Parameters
----------
W : ndarray, shape (n_features, n_features)
Input matrix
A : ndarray, shape (n_features, n_features)
Input matrix
Returns
-------
d : float
The Amari distance
"""
P = np.dot(W, A)
def s(r):
return np.sum(np.sum(r ** 2, axis=1) / np.max(r ** 2, axis=1) - 1)
return (s(np.abs(P)) + s(np.abs(P.T))) / (2 * P.shape[0])
def one_expe(n, p, sigma, bw, n_samples):
W = sigma * np.random.randn(p, p)
A = np.linalg.pinv(W)
S = np.random.laplace(size=(p, n))
X = np.dot(A, S)
X = torch.tensor(X, dtype=torch.float)
def score(w):
N, _ = w.shape
w_list = w.reshape(N, p, p)
z = w_list.matmul(X)
psi = torch.tanh(z).matmul(X.t()) / n - torch.inverse(
w_list
).transpose(-1, -2)
sc = -psi - w_list / sigma
return sc.reshape(N, p ** 2)
x = torch.randn(n_samples, p ** 2)
x_final = ksdd_lbfgs(x.clone(), score, bw=bw)
x_svgd = svgd(x.clone(), score, 0.1, bw=bw, max_iter=3000)
score_svgd = torch.norm(score(x_svgd)).item()
score_final = torch.norm(score(x_final)).item()
score_random = torch.norm(score(x)).item()
w_list = (x_final.reshape(n_samples, p, p)).detach().numpy()
w_svgd = (x_svgd.reshape(n_samples, p, p)).detach().numpy()
amari_ksd = np.sort([amari_distance(w, A) for w in w_list])
amari_svgd = np.sort([amari_distance(w, A) for w in w_svgd])
amari_random = np.sort(
[amari_distance(np.random.randn(p, p), A) for w in w_svgd]
)
return (
amari_ksd,
amari_svgd,
amari_random,
score_final,
score_svgd,
score_random,
)
p_list = [2]
n = 1000
sigma = 1
bw = 0.1
n_samples = 10
n_tries = 3
d_save = {}
for p in p_list:
print(p)
d_save[p] = {}
amari_ksds = []
amari_svgds = []
amari_randoms = []
score_ksds = []
score_svgds = []
score_randoms = []
for i in range(n_tries):
(
amari_ksd,
amari_svgd,
amari_random,
score_ksd,
score_svgd,
score_random,
) = one_expe(n, p, sigma, bw, n_samples)
amari_ksds.append(amari_ksd)
amari_svgds.append(amari_svgd)
amari_randoms.append(amari_random)
score_ksds.append(score_ksd)
score_svgds.append(score_svgd)
score_randoms.append(score_random)
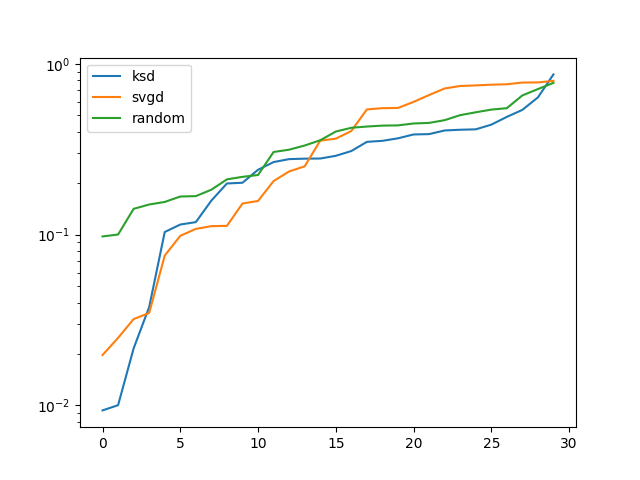
plt.figure()
plt.plot(np.sort(np.ravel(amari_ksds)), label="ksd")
plt.plot(np.sort(np.ravel(amari_svgds)), label="svgd")
plt.plot(np.sort(np.ravel(amari_randoms)), label="random")
plt.yscale("log")
plt.legend()
plt.show()
Total running time of the script: ( 0 minutes 38.349 seconds)