Note
Click here to download the full example code
Comparing Picard and FastICA for the task of detecting ECG artifacts in MEG¶
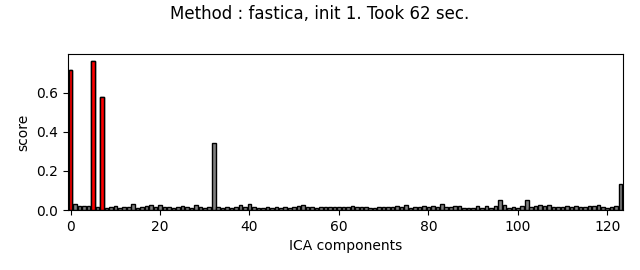
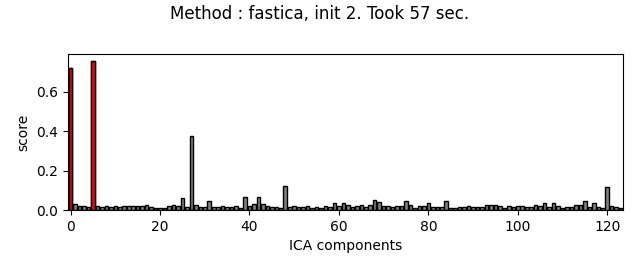
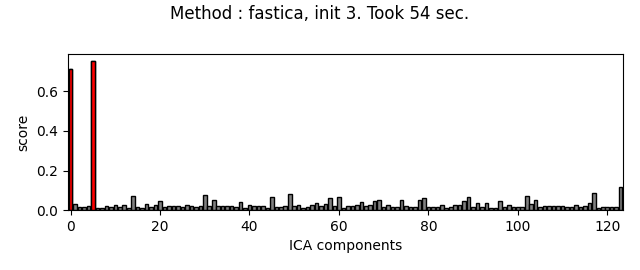
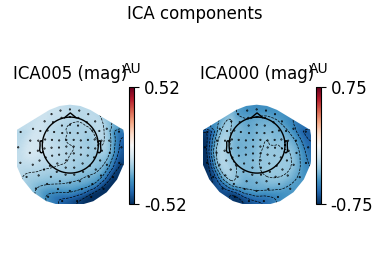
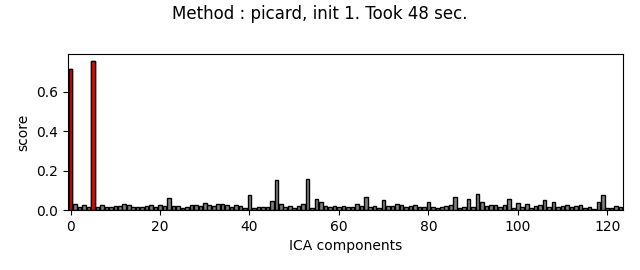
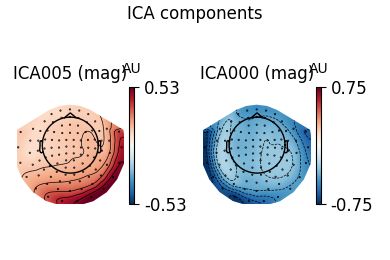
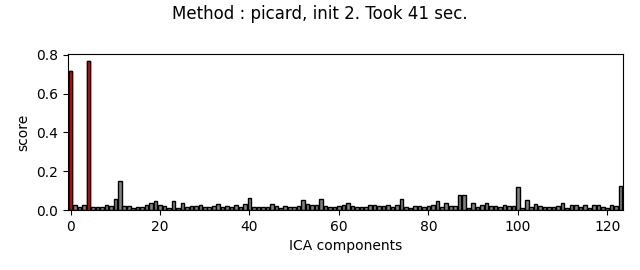
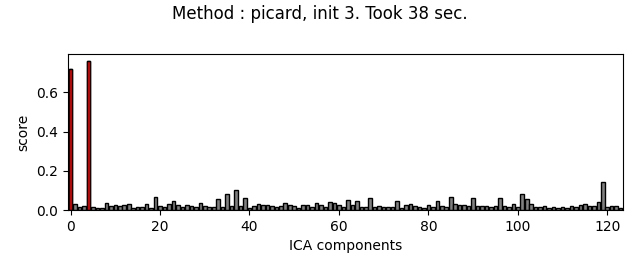
Picard and FastICA are fitted to MEG data, from several initializations. The scores related to the detection of ECG artifacts are displayed for each run. Picard is faster, and less dependent on the initialization.
# Authors: Pierre Ablin <pierreablin@gmail.com>
# Alexandre Gramfort <alexandre.gramfort@telecom-paristech.fr>
#
# License: BSD (3-clause)
from time import time
import mne
from mne.preprocessing import ICA
from mne.preprocessing import create_ecg_epochs
from mne.datasets import sample
Setup paths and prepare raw data.
mne.set_log_level(verbose='CRITICAL')
data_path = sample.data_path()
raw_fname = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw.fif'
raw = mne.io.read_raw_fif(raw_fname, preload=True)
raw.filter(1, None, fir_design='firwin') # already lowpassed @ 40
raw.set_annotations(mne.Annotations([1], [10], 'BAD'))
# raw.plot(block=True)
# For the sake of example we annotate first 10 seconds of the recording as
# 'BAD'. This part of data is excluded from the ICA decomposition by default.
# To turn this behavior off, pass ``reject_by_annotation=False`` to
# :meth:`mne.preprocessing.ICA.fit`.
raw.set_annotations(mne.Annotations([0], [10], 'BAD'))
picks = mne.pick_types(raw.info, meg=True, eeg=False, eog=False,
stim=False, exclude='bads')
Define a function that fits ICA to the data, identifies bad components related to ECG, and plots the resulting scores and topographies.
def fit_ica_and_plot_scores(method, random_state, tol=1e-4):
ica = ICA(n_components=0.95, method=method, random_state=random_state,
fit_params={'tol': tol}, max_iter=400)
t0 = time()
ica.fit(raw.copy(), picks=picks, decim=3,
reject=dict(mag=4e-12, grad=4000e-13), verbose='warning')
fit_time = time() - t0
title = ('Method : %s, init %d. Took %.2g sec.'
% (method, random_state + 1, fit_time))
ecg_epochs = create_ecg_epochs(raw, tmin=-.5, tmax=.5, picks=picks)
ecg_inds, scores = ica.find_bads_ecg(ecg_epochs, method='ctps')
ica.plot_scores(scores, exclude=ecg_inds, title=title)
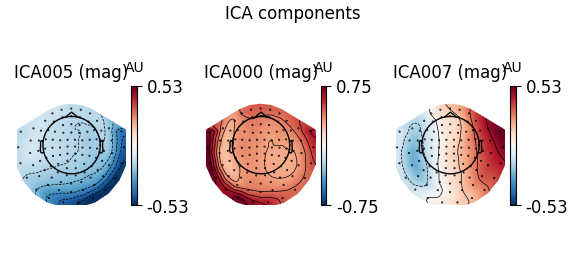
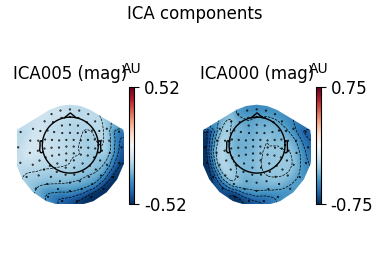
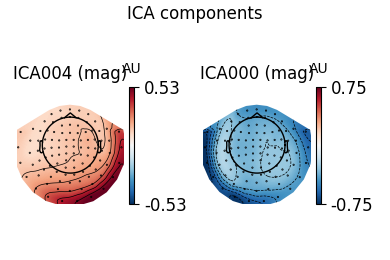
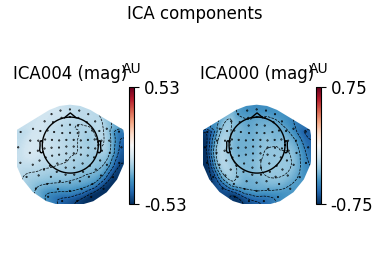
ica.plot_components(ecg_inds, colorbar=True)
Fit ICA using FastICA, with a few iterations, for different initializations :
n_inits = 3
method = 'fastica'
for random_state in range(n_inits):
fit_ica_and_plot_scores(method, random_state)
/home/circleci/.local/lib/python3.8/site-packages/sklearn/decomposition/_fastica.py:120: ConvergenceWarning: FastICA did not converge. Consider increasing tolerance or the maximum number of iterations.
warnings.warn(
/home/circleci/.local/lib/python3.8/site-packages/sklearn/decomposition/_fastica.py:120: ConvergenceWarning: FastICA did not converge. Consider increasing tolerance or the maximum number of iterations.
warnings.warn(
/home/circleci/.local/lib/python3.8/site-packages/sklearn/decomposition/_fastica.py:120: ConvergenceWarning: FastICA did not converge. Consider increasing tolerance or the maximum number of iterations.
warnings.warn(
Do the same thing with Picard :
method = 'picard'
for random_state in range(n_inits):
fit_ica_and_plot_scores(method, random_state)
The third topography found by FastICA does not really look like an ECG artifact, and is not consistent across different initializations. Picard finds the same topographies each time, and more consistently.
Total running time of the script: ( 5 minutes 20.973 seconds)